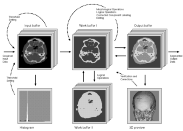
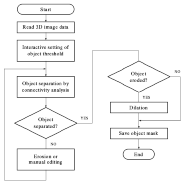
Our segmentation program ISEG provides the operator with a simple set of segmentation tools such as thresholding, connected component labeling, manual editing and morphologic and logic operations. As to which of them are used, and in which order, depends on the operator's goals, knowledge and experience.
The main advantage of this interactive approach is its independence of the scanning modality and the scanner parameter setting, since the actual sequence of operations and their parameters are selected based on the data properties and the desired goal. Further, the operator gets a feeling of the fidelity of the segmentation results and subseqently mrendered image, due to the loop back provided by the program.
The technique originates from a simple presumption that the object represents an approximately homogeneous region, which differs from other regions by the density level. Thresholding is therefore the basic segmentation operation.
Since different tissues can have densities with overlapping intervals, a situation is possible, when more than the desired object is marked. Such regions can be separated on presumption of object connectivity.
However, the object separation can be unsuccessful.
Such an object either really touches other objects or tissues,
or the connection is mediated by
some inhomogeneity or small object.
It is known that morphological operation of opening
(erosion followed by dilation) can delete thin connections between
objects.
This ability has proven to be not effective enough for our purposes.
Therefore an additional operation of connected component labeling
has been inserted between these two operations.
The interactive segmentation system ISEG offers the operator a set of simple segmentation tools, which enable the user to reach the desired goal on the basis of his knowledge. Which of the offered tools would be used and in which order, depends on the given goal, the quality of the data set (noise, blurring) and, of course, also on the users skills. In the following a list of possible procedures is presented, which turned out to be useful in practice.
Let us denote the single operations as follows: T - thresholding E - erosion D - dilation C - connected component labeling M - manual editing I - inversion. (X) represents result of the operation X stored in a buffer and (X) & (Y) represents (binary AND) operation between two buffers.
The following sequences were found to be useful:
| T | Simple thresholding |
| TC | Separation of objects |
| TECD, TMC | Separation of objects with interconnections |
| (TE(1)CD(n)) & (T) | The initial erosion is repeated once, dilations are repeated n-times. AND-ing the segmented object with the original mask minimizes distortion introduced by multiple dilations |
| (T1ECD) & (T2) | Thresholding by T1, defining a narrower interval of gray values than the correct threshold T2, can help in object separation. AND-ing with the correct mask minimize the distortion again |
| TICI | Filling holes in the object |
The ISEG program has been implemented on an HP 9000/720 workstation under the UNIX operating system. The graphical subsystem is based on the X Window System X11R5 and OSF Motif version 1.2; written in C and User Interface Language (UIL).
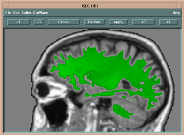
The backbone of the segmentation program is composed of three 3D buffers (see the following Figure):
 |
 |
| Block diagram of the ISEG program | Manual Edit Window |
 |
 |
| Main window | Render Window |
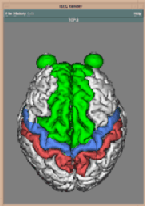
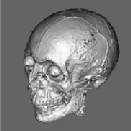
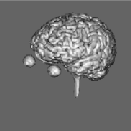
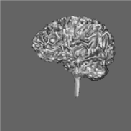
Segmentation of data for binary volume rendering has been the basic goal of the development of the whole system. The following figures show an example of an MRI data set (human head), segmented into 15 objects, such as skin, left and right brain hemispheres, cerebellum, brain stem, ventricles, etc.
 |
 |
 |
 |
It became apparent, that the ISEG system can be advantageously used also for improvement of the performance and scope of application of various probabilistic volume rendering methods. In this case all scene voxels can contribute to the final image.
It is assumed that distinct tissues occupy distinct regions in the gray level histogram of the scene and that spatially each tissue touches only two neighbors at most: one which is in the histogram adjacent from the left and one from the right side. These criteria allow to assign some opacity to each tissue with a piecewise linear mapping in regions, which cannot be unambiguously assigned to one of the tissues. This segmentation approach improves visualization of poorly defined boundaries, small details, and thin structures in respect to binary methods based on all-or-nothing decisions.
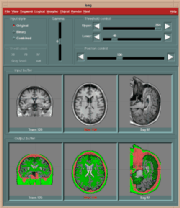
However, the strong demands of the model limit the usefulness of the volume rendering approach only to a small class of data. For example, the CT data with four main tissues (air, skin, soft tissue and bone) satisfy the model, while MR scans as a whole do not. However, in such data sets it is possible to define interactively regions of interest, where the volume rendering model conditions are met and thus enable to take advantage of this kind of visualization approach. The following figure shows an example of a brain ventricular system (MR data) rendered by a surface as well as a volume rendering approach. The mask (b) follows the surface of the brain white matter and thereby selects only white matter, ventricles and small structures around ventricles (nucleus caudatus, putamen). Comparing the images (c) and (d) we can see the differences between the binary and probabilistic volume rendering approaches. In this case, the (d) image shows details with higher fidelity than the surface rendered image (c) does.
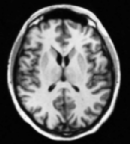 |
 |
 |
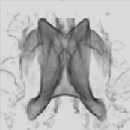 |
| (a) | (b) | (c) | (d) |
| Interactive segmentation for volume rendering: (a) original MR slice, (b) interactively defined mask, (c) surface and (d) volume rendered projection. | |||
|---|---|---|---|
(Full list of publications)
|
Back to Milos Sramek's home page |
|